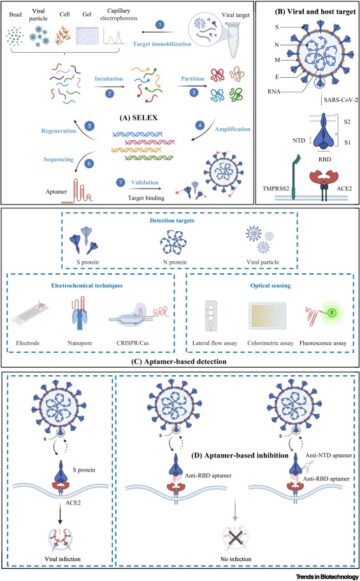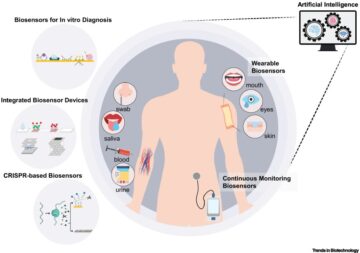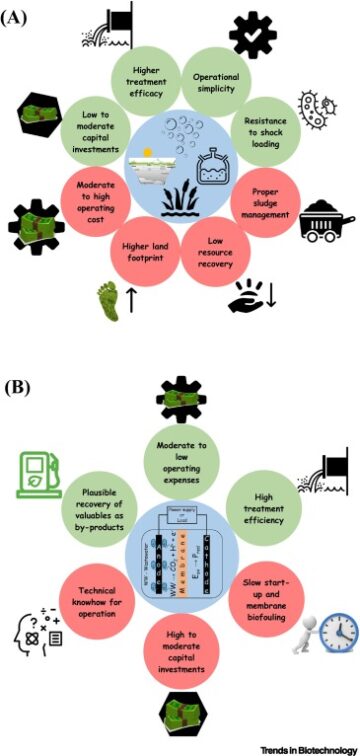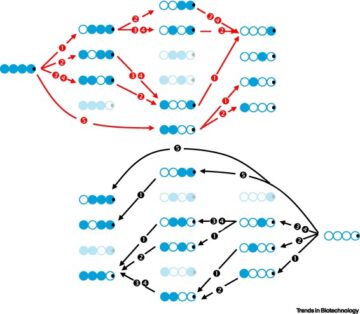
Drugging membrane protein interactions.
Annu. Rev. Biomed. Eng. 2016; 18: 51-76
Cryo-electron microscopy: moving beyond X-ray crystal structures for drug receptors and drug development.
Annu. Rev. Pharmacol. Toxicol. 2019; 60: 51-71
Surface plasmon resonance spectroscopy for characterisation of membrane protein–ligand interactions and its potential for drug discovery.
Biochim. Biophys. Acta. 2014; 1838: 43-55
Structure determination of membrane proteins by NMR spectroscopy.
Chem. Rev. 2004; 104: 3587-3606
Overcoming the challenges of membrane protein crystallography.
Curr. Opin. Struct. Biol. 2008; 18: 581-586
Detergents for the stabilization and crystallization of membrane proteins.
Methods. 2007; 41: 388-397
Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers.
Prot. Sci. 2009; 12: 2476-2481
Functional reconstitution of β2-adrenergic receptors utilizing self-assembling nanodisc technology.
Biotechniques. 2006; 40: 601-612
Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size.
J. Am. Chem. Soc. 2004; 126: 3477-3487
Phospholipid phase transitions in homogeneous nanometer scale bilayer discs.
FEBS Lett. 2004; 556: 260-264
Functional and structural comparison of the ABC exporter MsbA studied in detergent and reconstituted in nanodiscs.
Biochem. Biophys. Res. Commun. 2019; 512: 448-452
Probing membrane protein assembly into nanodiscs by in situ dynamic light scattering: A2a receptor as a case study.
Biology (Basel). 2020; 9: 400
Real-time monitoring of binding events on a thermostabilized human A2A receptor embedded in a lipid bilayer by surface plasmon resonance.
BBA Biomembr. 2015; 1848: 1224-1233
Structure of the M2 muscarinic receptor–β-arrestin complex in a lipid nanodisc.
Nature. 2020; 579: 297-302
The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs.
Cell. 2016; 167: 763-773
Reconstitution and functional characterization of ion channels from nanodiscs in lipid bilayers.
J. Gen. Physiol. 2018; 150: 637-646
Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science. 2018; 359: 228-232
Nanodiscs for immobilization of lipid bilayers and membrane receptors: kinetic analysis of cholera toxin binding to a glycolipid receptor.
Anal. Chem. 2008; 80: 6245-6252
Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties.
PNAS. 2006; 103: 11509-11514
A saposin–lipoprotein nanoparticle system for membrane proteins.
Nat. Methods. 2016; 13: 345-351
Saposin lipid nanoparticles: a highly versatile and modular tool for membrane protein research.
Structure. 2018; 26: 345-355
The styrene–maleic acid copolymer: a versatile tool in membrane research.
Eur. Biophys. J. 2016; 45: 3-21
Polymer nanodiscs: advantages and limitations.
Chem. Phys. Lipids. 2019; 219: 45-49
Polymer nanodiscs: discoidal amphiphilic block copolymer membranes as a new platform for membrane proteins.
Sci. Rep. 2017; 7: 15227
Lipid nanodiscs via ordered copolymers.
Chem. 2020; 6: 2782-2795
Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum.
Science. 2016; 354: 890-893
Development of methodology to investigate the surface SMALPome of mammalian cells.
Front. Mol. Biosci. 2021; 8780033
Production of human A2AAR in lipid nanodiscs for 19F-NMR and single-molecule fluorescence spectroscopy.
STAR Protoc. 2022; 3101535
Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR.
Nat. Protoc. 2018; 13: 79-98
DirectMX – One-step reconstitution of membrane proteins from crude cell membranes into salipro nanoparticles.
Front. Bioeng. Biotechnol. 2020; 8: 215
Saposin–lipoprotein scaffolds for structure determination of membrane transporters.
Methods Enzymol. 2017; 594: 85-99
Influence of DIBMA polymer length on lipid nanodisc formation and membrane protein extraction.
Biomacromolecules. 2021; 22: 763-772
Detergent-free isolation of functional G protein-coupled receptors into nanometric lipid particles.
Biochemistry. 2016; 55: 38-48
Detergent-free solubilisation & purification of a G protein coupled receptor using a polymethacrylate polymer.
Biochim. Biophys. Acta Biomembr. 2021; 1863183441
Detergent alternatives: membrane protein purification using synthetic nanodisc polymers.
Methods Mol. Biol. 2022; 2507: 375-387
Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins.
Nano Lett. 2002; 2: 853-856
Biophysical characterization of membrane proteins in nanodiscs.
Methods. 2013; 59: 287-300
The charge properties of phospholipid nanodiscs.
Biophys. J. 2016; 111: 989-998
Rapid preparation of nanodiscs for biophysical studies.
Arch. Biochem. Biophys. 2021; 712109051
Express incorporation of membrane proteins from various human cell types into phospholipid bilayer nanodiscs.
Biochem. J. 2017; 474: 1361-1371
Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers.
Biotechniques. 2003; 35: 556-563
Co-incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers.
Arch. Biochem. Biophys. 2004; 424: 141-153
Nanodisc-solubilized membrane protein library reflects the membrane proteome.
Anal. Bioanal. Chem. 2013; 405: 4009-4016
Cryo-EM structure of the native rhodopsin dimer in nanodiscs.
J. Biol. Chem. 2019; 294: 14215-14230
Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins.
J. Biol. Chem. 2007; 282: 14875-14881
Structure of the neurotensin receptor 1 in complex with β-arrestin 1.
Nature. 2020; 579: 303-308
Molecular basis of β-arrestin coupling to formoterol-bound β1-adrenoceptor.
Nature. 2020; 583: 862-866
Emerging structural insights into GPCR–β-arrestin interaction and functional outcomes.
Curr. Opin. Struct. Biol. 2022; 75102406
Cryo-EM structure of an activated GPCR–G protein complex in lipid nanodiscs.
Nat. Struct. Mol. Biol. 2021; 28: 258-267
Covalently circularized nanodiscs for studying membrane proteins and viral entry.
Nat. Methods. 2017; 14: 49-52
Circularized and solubility-enhanced MSPs facilitate simple and high-yield production of stable nanodiscs for studies of membrane proteins in solution.
FEBS J. 2019; 286: 1734-1751
Nanodiscs incorporating native β1 adrenergic receptor as a novel approach for the detection of pathological autoantibodies in patients with dilated cardiomyopathy.
J. Appl. Lab. Med. 2019; 4: 391-403
Nanodisc technology facilitates identification of monoclonal antibodies targeting multi-pass membrane proteins.
Sci. Rep. 2020; 10: 1130
A tetrameric assembly of saposin A: increasing structural diversity in lipid transfer proteins.
Contact. 2021; 4: 1-11
Saposins: structure, function, distribution, and molecular genetics.
J. Lipid Res. 1992; 33: 1255-1267
Structure of saposin A lipoprotein discs.
PNAS. 2011; 109: 2908-2912
An adaptable phospholipid membrane mimetic system for solution NMR studies of membrane proteins.
J. Am. Chem. Soc. 2017; 139: 14829-14832
Purification of a native nicotinic receptor.
Methods Enzymol. 2021; 653: 189-206
Footprinting mass spectrometry of membrane proteins: ferroportin reconstituted in saposin A picodiscs.
Anal. Chem. 2021; 93: 11370-11378
Interactions of a bacterial RND transporter with a transmembrane small protein in a lipid environment.
Structure. 2020; 28: 625-634
Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat. Commun. 2022; 13: 4366
Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins.
Neuron. 2020; 106: 952-962
Structure and gating mechanism of the α7 nicotinic acetylcholine receptor.
Cell. 2021; 184: 2121-2134
Agonist selectivity and Ion permeation in the α3β4 ganglionic nicotinic receptor.
Neuron. 2019; 104: 501-511
Membrane protein extraction and purification using partially-esterified SMA polymers.
Biochim. Biophys. Acta Biomembr. 2021; 1863183758
Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer.
J. Am. Chem. Soc. 2009; 131: 7484-7485
Detergent-free extraction of a functional low-expressing GPCR from a human cell line.
Biochim. Biophys. Acta Biomembr. 2020; 1862183152
Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure.
Biochem. J. 2016; 473: 4349-4360
Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs.
Proc. Natl. Acad. Sci. U. S. A. 2014; 111: 18607-18612
Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights.
Methods. 2018; 147: 106-117
Structure and activity of lipid bilayer within a membrane-protein transporter.
PNAS. 2018; 115: 12985-12990
Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure.
Biochim. Biophys. Acta Biomembr. 2018; 1860: 378-383
Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc. Natl. Acad. Sci. U. S. A. 2021; 118e2106750118
Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature. 2018; 557: 123-126
An acid-compatible co-polymer for the solubilization of membranes and proteins into lipid bilayer-containing nanoparticles.
Nanoscale. 2018; 10: 10609-10619
Bioinspired, size-tunable self-assembly of polymer–lipid bilayer nanodiscs.
Angew. Chem. Int. Ed. 2017; 56: 11466-11470
Formation of pH-resistant monodispersed polymer–lipid nanodiscs.
Angew. Chem. 2018; 130: 1356-1359
Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs.
Chem. Commun. 2018; 54: 9615-9618
Measurement of residual dipolar couplings using magnetically aligned and flipped nanodiscs.
Langmuir. 2022; 38: 244-252
Magnetic alignment of polymer macro-nanodiscs enables residual-dipolar-coupling-based high-resolution structural studies by NMR spectroscopy.
Angew. Chem. Int. Ed. 2019; 58: 14925-14928
Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer.
Angew. Chem. 2017; 56: 1919-1924
Spontaneous lipid nanodisc fomation by amphiphilic polymethacrylate copolymers.
J. Am. Chem. Soc. 2017; 139: 18657-18663
Synthesis, characterization, and nanodisc formation of non-ionic polymers.
Angew. Chem. Int. Ed. 2021; 60: 16885-16888
A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification.
Biochim. Biophys. Acta Biomembr. 2020; 1862183281
Lipid dynamics in diisobutylene-maleic acid (DIBMA) lipid particles in presence of sensory rhodopsin II.
Int. J. Mol. Sci. 2021; 22: 2548
Functional solubilization of the β2-adrenoceptor using diisobutylene maleic acid.
iScience. 2021; 24103362
Formation of lipid-bilayer nanodiscs by diisobutylene/maleic acid (DIBMA) copolymer.
Langmuir. 2017; 33: 14378-14388
Enhancing the stability and homogeneity of non-ionic polymer nanodiscs by tuning electrostatic interactions.
J. Colloid Interface Sci. 2023; 634: 887-896
Non-ionic inulin-based polymer nanodiscs enable functional reconstitution of a redox complex composed of oppositely charged CYP450 and CPR in a lipid bilayer membrane.
Anal. Chem. 2022; 94: 11908-11915
Comparison of lipidic carrier systems for integral membrane proteins – MsbA as case study.
Biol. Chem. 2019; 400: 1509-1518
Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation.
J. Biol. Chem. 2007; 282: 7066-7076
Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers.
Protein Eng. Des. Sel. 2010; 23: 843-848
Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J. Am. Chem. Soc. 2013; 135: 1919-1925
A split-intein-based method for the efficient production of circularized nanodiscs for structural studies of membrane proteins.
ChemBioChem. 2018; 19: 1927-1933
Optimization of the production of covalently circularized nanodiscs and their characterization in physiological conditions.
Langmuir. 2018; 34: 3525-3532
One-step construction of circularized nanodiscs using SpyCatcher-SpyTag.
Nat. Commun. 2021; 12: 5451
- SEO Powered Content & PR Distribution. Get Amplified Today.
- Platoblockchain. Web3 Metaverse Intelligence. Knowledge Amplified. Access Here.
- Source: https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(23)00057-4?rss=yes
- ][p
- 1
- a
- ABC
- Activation
- activity
- advances
- advantages
- AL
- aligned
- alternative
- alternatives
- analysis
- and
- Antibodies
- approach
- AS
- Assembly
- authors
- Basel
- basis
- Beyond
- binding
- Block
- bound
- by
- case
- case study
- Cells
- challenges
- Channel
- channels
- charge
- charged
- comparison
- complex
- composed
- Compound
- conditions
- construction
- controlled
- conventional
- coupled
- crude
- Crystal
- Defense
- Detection
- detergents
- determination
- Determine
- Development
- discovery
- Disease
- distribution
- Diversity
- drug
- drug development
- drug discovery
- dynamic
- dynamics
- e
- ed
- effect
- efficient
- embedded
- enable
- enables
- entry
- Environment
- Ether (ETH)
- events
- expressed
- extraction
- facilitate
- facilitates
- For
- formation
- Free
- from
- function
- functional
- Gen
- Genetics
- high-resolution
- highly
- HTTPS
- human
- i
- Identification
- in
- incorporating
- increasing
- insights
- integral
- interaction
- interactions
- Interface
- investigate
- isolation
- ITS
- jpg
- kidney
- lab
- Length
- Library
- light
- limitations
- Line
- List
- M2
- Mass
- mechanism
- method
- Methodology
- methods
- Microscopy
- modular
- MOL
- molecular
- monitoring
- moving
- Multi-pass
- native
- New
- New Platform
- novel
- of
- on
- ONE
- opportunities
- particle
- patients
- phase
- platform
- Platforms
- plato
- Plato Data Intelligence
- PlatoData
- polymer
- Polymers
- potential
- power
- presence
- Product
- Production
- properties
- Protein
- Proteins
- reflects
- research
- resonance
- reveal
- s
- Scale
- SCI
- separate
- Simple
- single
- site
- Size
- SMA
- small
- solution
- Spectroscopy
- Spin
- Stability
- stable
- State
- States
- strategies
- structural
- structure
- studied
- studies
- Study
- Studying
- Surface
- synthetic
- system
- Systems
- targeting
- Technology
- The
- their
- to
- tool
- transfer
- transitions
- types
- Utilizing
- various
- versatile
- via
- W
- with
- within
- x-ray
- zephyrnet