Lead Inventors
Advantages
- DNA as building blocks
- Broad applications
- Low cost with big potential

DNA nanostructures with their potential for cell and tissue permeability, biocompatibility, and high programmability at the nanoscale level are promising candidates as new types of drug delivery vehicles, highly specific diagnostic devices, and tools to decipher how biomolecules dynamically change their shapes, and interact with each other and with candidate drugs. Wyss Institute researchers are providing a suite of diverse, multifunctional DNA nanotechnological tools with unique capabilities and potential for a broad range of clinical and biomedical research areas.
DNA nanotechnological devices for therapeutic drug delivery
DNA nanostructures have future potential to be widely used to transport and present a variety of biologically active molecules such as drugs and immune-enhancing antigens and adjuvants to target cells and tissues in the human body.
DNA origami as high-precision delivery components of cancer vaccines
The Wyss Institute has developed cancer vaccines to improve immunotherapies. These approaches use implantable or injectable biomaterial-based scaffolds that present tumor-specific antigens, and biomolecules that attract dendritic immune cells (DCs) into the scaffold, and activate them so that after their release they can orchestrate anti-tumor T cell responses against tumors carrying the same antigens. To be activated most effectively, DCs likely need to experience tumor antigens and immune-boosting CpG adjuvant molecules at particular ratios (stoichiometries) and configurations that register with the density and distribution of receptor molecules on their cell surface.
Specifically developed DNA origami, programmed to assemble into rigid square-lattice blocks that co-present tumor antigens and adjuvants to DCs within biomaterial scaffolds with nanoscale precision have the potential to boost the efficacy of therapeutic cancer vaccines, and can be further functionalized with anti-cancer drugs.
Chemical modification strategy to protect drug-delivering DNA nanostructures
DNA nanostructures such as self-assembling DNA origami are promising vehicles for the delivery of drugs and diagnostics. They can be flexibly functionalized with small molecule and protein drugs, as well as features that facilitate their delivery to specific target cells and tissues. However, their potential is hampered by their limited stability in the body’s tissues and blood. To help fulfill the extraordinary promise of DNA nanostructures, Wyss researchers developed an easy, effective and scalable chemical cross-linking approach that can provide DNA nanostructures with the stability they need as effective vehicles for drugs and diagnostics.
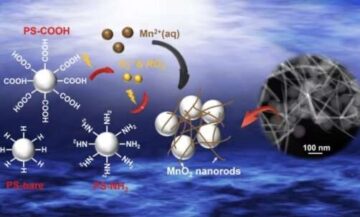
In two simple cost-effective steps, the Wyss’ approach first uses a small-molecule, unobtrusive neutralizing agent, PEG-oligolysine, that carries multiple positive charges, to cover DNA origami structures. In contrast to commonly used Mg2+ions that each neutralize only two negative changes in DNA structures, PEG-oligolysine covers multiple negative charges at one, thus forming a stable “electrostatic net,” which increases the stability of DNA nanostructures about 400-fold. Then, by applying a chemical cross-linking reagent known as glutaraldehyde, additional stabilizing bonds are introduced into the electrostatic net, which increases the stability of DNA nanostructures by another 250-fold, extending their half-life into a range that is compatible with a broad range of clinical applications.
DNA nanotechnological devices as ultrasensitive diagnostic and analytical tools
The generation of detectable DNA nanostructures in response to a disease or pathogen-specific nucleic acids, in principle, offers a means for highly effective biomarker detection in diverse samples. A single molecule binding event of a synthetic oligonucleotide to a target nucleic acid can nucleate the creation of much larger structures by the cooperative assembly of smaller synthetic DNA units like DNA tiles or bricks into larger structures that then can be visualized in simple laboratory assays. However, a central obstacle to these approaches is the occurrence of (1) non-specific binding and (2) non-specific nucleation events in the absence of a specific target nucleic acid which can lead to false-positive results. Wyss DNA nanotechnologists have developed two separately applicable but combinable solutions for these problems.
Digital counting of biomarker molecules with DNA nanoswitch catenanes
To enable the initial detection (binding) of biomarkers with ultra-high sensitivity and specificity, Wyss researchers have developed a type of DNA nanoswitch that, designed as a larger catenane (Latin catenameaning chain), is assembled from mechanically interlocked ring-shaped substructures with specific functionalities that together enable the detection and counting of single biomarker molecules. In the “DNA Nanoswitch Catenane” structure, both ends of a longer synthetic DNA strand are linked to two antibody fragments that each specifically bind different parts of the same biomarker molecule of interest, thus allowing for high target specificity and sensitivity.
This bridging-event causes the strand to close into a “host ring,” which it is interlocked at different regions with different “guest rings.” Closing of the host ring switches the guest rings into a configuration that allows the synthesis of a new DNA strand. The newly synthesized diagnostic strand then can be unambiguously detected as a single digital molecule count, while disrupting the antibody fragment/biomarker complex starts a new biomarker counting cycle. Both, the target binding specificity and the synthesis of a target-specific DNA strand also enable the combination of multiple DNA nanoswitch catenanes to simultaneously count different biomarker molecules in a single multiplexed reaction.
For ultrasensitive diagnostics, it is desirable to have the fastest amplification and the lowest rate of spurious nucleation. DNA nanotechnology approaches have the potential to deliver this in an enzyme-free, low-cost manner.
WILLIAM SHIH
A rapid amplification platform for diverse biomarkers
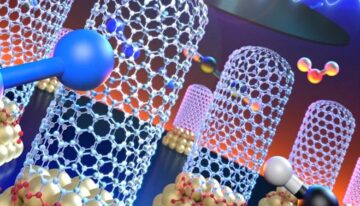
A rapid, low-cost and enzyme-free detection and amplification platform avoids non-specific nucleation and amplification and allows the self-assembly of much larger micron-scale structures from a single seed in just minutes. The method, called “Crisscross Nanoseed Detection” enables the ultra-cooperative assembly of ribbons starting from a single biomarker binding event. The micron-scale structures are densely woven from single-stranded “DNA slats,” whereby an inbound slat snakes over and under six or more previously captured slats on a growing ribbon end in a “crisscross” manner, forming weak but highly-specific interactions with its interacting DNA slats. The nucleation of the assembly process is strictly target-seed specific and the assembly can be carried out in a one-step reaction in about 15 minutes without the addition of further reagents, and over a broad range of temperatures. Using standard laboratory equipment, the assembled structures then can be rapidly visualized or otherwise detected, for example, using high-throughput fluorescence plate reader assays.
CURRENT OPPORTUNITY – STARTUP
Crisscross Nanoseed Detection: Nanotechnology-Powered Infectious Disease Diagnostics
Enzyme-free DNA nanotechnology for rapid, ultrasensitive, and low-cost detection of infectious disease biomarkers with broad accessibility in point-of-care settings.
The DNA assembly process in the Crisscross Nanoseed Detection method can also be linked to the action of DNA nanoswitch catenanes that highly specifically detect a biomarker molecule leading to preservation of a molecular record. Each surviving record can nucleate the assembly of a crisscross nanostructure, combining high-specificity binding with amplification for biomarker detection.
Wyss researchers are currently developing the approach as a multiplexable low-cost diagnostic for the COVID-19 causing SARS-CoV-2 virus and other pathogens that could give accurate results faster and at lower costs than currently used techniques.
Nanoscale devices for determining the structure and identity of proteins at the single-molecule level
The ability to identify and quantify proteins from trace biological samples would have a profound impact on both basic research and clinical practice, from monitoring changes in protein expression within individual cells, to enabling the discovery of new biomarkers of disease. Furthermore, the ability to also determine their structures and interactions would open up new avenues for drug discovery and characterization. Over the past decades, developments in DNA analysis and sequencing have unquestionably revolutionized medicine – yet equivalent developments for protein analysis have remained a challenge. While methods such as mass spectrometry for protein identification, and cryoEM for structure determination have rapidly advanced, challenges remain regarding resolution and the ability to work with trace heterogeneous samples.
To help meet this challenge, researchers at the Wyss Institute have developed a new approach that combines DNA nanotechnology with single-molecule manipulation to enable the structural identification and analysis of proteins and other macromolecules. “DNA Nanoswitch Calipers” (DNCs) offer a high-resolution approach to “fingerprint proteins” by measuring distances and determining geometries within single proteins in solution. DNCs are nanodevices designed to measure distances between DNA handles that have been attached to target molecules of interest. DNC states can be actuated and read out using single-molecule force spectroscopy, enabling multiple absolute distance measurements to be made on each single-molecule.
DNCs could be widely adapted to advance research in different areas, including structural biology, proteomics, diagnostics and drug discovery.
All technologies are in development and available for industry collaborations.
.wordads-ad-wrapper { display:none; font: normal 11px Arial, sans-serif; letter-spacing: 1px; text-decoration: none; width: 100%; margin: 25px auto; padding: 0; } .wordads-ad-title { margin-bottom: 5px; } .wordads-ad-controls { margin-top: 5px; text-align: right; } .wordads-ad-controls span { cursor: pointer; } .wordads-ad { width: fit-content; margin: 0 auto; }
- SEO Powered Content & PR Distribution. Get Amplified Today.
- Platoblockchain. Web3 Metaverse Intelligence. Knowledge Amplified. Access Here.
- Source: https://genesisnanotech.wordpress.com/2023/01/16/dna-nanotechnology-tools-from-design-to-applications-current-opportunities-and-collaborations-wyss-institute-harvard-university/
- 1
- 11
- a
- ability
- About
- Absolute
- accessibility
- accurate
- Action
- active
- adapted
- addition
- Additional
- advance
- advanced
- After
- against
- Allowing
- allows
- analysis
- Analytical
- and
- Another
- applicable
- applications
- Applying
- approach
- approaches
- areas
- assembled
- Assembly
- auto
- available
- basic
- between
- Big
- bind
- binding
- biology
- biomarker
- biomedical
- blood
- body
- Bonds
- boost
- broad
- Building
- called
- Cancer
- candidate
- candidates
- capabilities
- carrying
- causes
- causing
- Cells
- central
- chain
- challenge
- challenges
- change
- Changes
- charges
- chemical
- Clinical
- Close
- closing
- collaborations
- combination
- combines
- combining
- commonly
- compatible
- complex
- components
- Configuration
- contrast
- cooperative
- Cost
- cost-effective
- Costs
- could
- cover
- covers
- COVID-19
- cpg
- creation
- Current
- Currently
- decades
- Decipher
- deliver
- delivery
- Design
- designed
- detected
- Detection
- determination
- Determine
- determining
- developed
- developing
- Development
- developments
- Devices
- different
- digital
- discovery
- Disease
- Display
- distance
- distribution
- diverse
- dna
- drug
- drug discovery
- Drugs
- each
- Effective
- effectively
- enable
- enables
- enabling
- ends
- equipment
- Equivalent
- Event
- events
- example
- experience
- extending
- extraordinary
- facilitate
- faster
- fastest
- Features
- Files
- First
- Force
- from
- Fulfill
- functionalities
- further
- Furthermore
- future
- generation
- Give
- Growing
- Guest
- Handles
- harvard
- harvard university
- help
- High
- high-resolution
- highly
- host
- How
- However
- HTTPS
- human
- Identification
- identify
- Identity
- Impact
- improve
- in
- Including
- Increases
- individual
- industry
- initial
- Institute
- interact
- interacting
- interactions
- interest
- introduced
- IT
- known
- laboratory
- larger
- Latin
- lead
- leading
- Level
- likely
- Limited
- linked
- longer
- made
- Manipulation
- manner
- Margin
- Mass
- max-width
- means
- measure
- measurements
- measuring
- medicine
- Meet
- method
- methods
- minutes
- molecular
- molecule
- monitoring
- more
- most
- multiple
- nanotechnology
- Nature
- Need
- negative
- net
- New
- normal
- obstacle
- offer
- Offers
- ONE
- open
- opportunities
- Opportunity
- Other
- otherwise
- Overcome
- particular
- parts
- past
- platform
- plato
- Plato Data Intelligence
- PlatoData
- positive
- potential
- practice
- Precision
- present
- previously
- principle
- problems
- process
- programmed
- promise
- promising
- protect
- Protein
- Proteins
- provide
- providing
- range
- rapid
- rapidly
- Rate
- reaction
- Read
- Reader
- record
- regarding
- regions
- register
- release
- remain
- remained
- research
- researchers
- Resolution
- response
- Results
- revolutionized
- Ribbon
- rigid
- Ring
- same
- SARS-CoV-2
- scalable
- seed
- Sensitivity
- settings
- shapes
- Simple
- simultaneously
- single
- SIX
- small
- smaller
- So
- solution
- Solutions
- specific
- specifically
- specificity
- Stability
- stable
- standard
- Starting
- starts
- States
- Steps
- Strategy
- structural
- structure
- such
- suite
- Surface
- synthetic
- Target
- techniques
- Technologies
- The
- their
- Therapeutic
- tissues
- to
- together
- tools
- trace
- transport
- types
- under
- understanding
- unique
- units
- university
- use
- vaccines
- variety
- Vehicles
- virus
- which
- while
- widely
- within
- without
- WordPress
- Work
- would
- zephyrnet