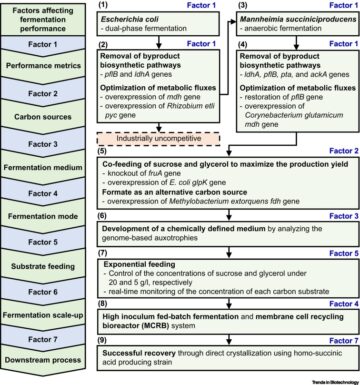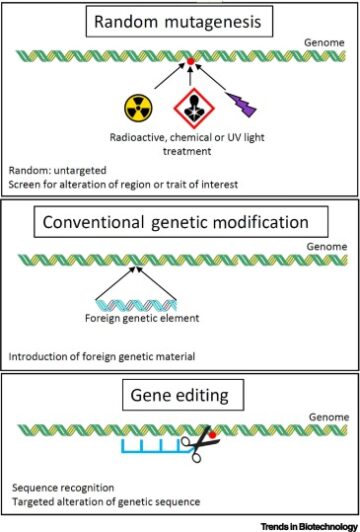
Glossary
Aptamer
an ssDNA or ssRNA oligonucleotide which binds its ligand, such as proteins or other NAs, with high specificity.
DNA helicase
a protein enzyme that unwinds dsDNA by ATP hydrolysis.
Extrachromosomal DNA
a form of dsDNA inside and outside the nucleus of a cell that does not belong to chromosomal DNA and has a circular shape.
Fluorescence in situ hybridization (FISH)
a cytogenetic technique in which fluorescently labeled probes hybridize with denatured genomic ssDNA in fixed cells or tissue samples. FISH is suitable for DNA localization studies and for uncovering genomic abnormalities ranging from numerical chromosomal changes to submicroscopic single-gene-level alterations.
Förster resonance energy transfer (FRET)
an energy transfer mechanism, over nanometer distance, from an excited donor fluorophore to an acceptor fluorophore as a consequence of resonance. Such donor-acceptor fluorophores are referred to as FRET pairs.
Intergenic DNA region
the noncoding DNA region that resides between two genes.
Mitochondrial DNA (mtDNA)
circularized DNA that resides in the mitochondria of eukaryotic cells. Human mtDNA encompasses 16.6 kb carrying 37 genes, of which 13 are protein coding and 24 are RNA genes. The copy number of mtDNA per cell can vary between 1000 and 100 000, depending on the cell type.
Molecular beacon (MB)
a single-stranded DNA molecule with a partially self-hybridized stem and a free loop structure. The distal ends of the beacon can be modified with a fluorophore and quencher (F/Q) pair. Under specific hybridization circumstances, the stem–loop structure will open up, thereby dissociating the F/Q pair.
Polycistronic vector
a vector that contains the genetic code for more than one gene. Polycistronic vectors produce a single mRNA molecule that leads to the expression of multiple proteins.
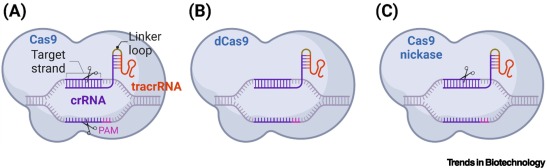
Protospacer-adjacent motif (PAM)
a Cas9-species-specific multinucleotide sequence (e.g., NGG for Sp. Cas9), positioned next to the nontarget strand, which is specifically recognized by the CRISPR-(d)Cas9 complex prior to binding.
Quantum dot (QD)
a nanoparticle with a specific crystal structure that possesses unique optical properties resulting from its small size.
Rolling circle amplification (RCA)
an NA amplification technique which is triggered by the hybridization of a single-stranded padlock probe to an NA target sequence of interest. The padlock probe is circularized by ligation and a primer is hybridized. DNA polymerase elongates the primer, creating a long linear product with thousands of copies of the padlock probe.
Single-nucleotide variant (SNV)
a general term for an alteration in a DNA sequence that involves the variation of a single nucleotide.
SNP
a type of single-nucleotide variation that occurs in germline genomic DNA and is present in at least 1% of the species’ population.
Telomere
a region of repetitive sequences that is situated at the end of linear chromosomes in eukaryotic cells. Telomeres protect the chromosomal DNA, enable complete replication of genetic material throughout the cell cycles, and can be involved in chromosome movement and positioning.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- Platoblockchain. Web3 Metaverse Intelligence. Knowledge Amplified. Access Here.
- Source: https://www.cell.com/trends/biotechnology/fulltext/S0167-7799(22)00274-8?rss=yes
- 000
- 100
- a
- and
- beacon
- between
- carrying
- Cells
- Changes
- Chromosomes
- Circle
- circumstances
- code
- Coding
- complete
- complex
- contains
- Creating
- Crystal
- cycles
- Depending
- distance
- dna
- DOT
- enable
- encompasses
- ends
- energy
- excited
- Fish
- fixed
- form
- Free
- from
- General
- generation
- High
- HTTPS
- human
- Imaging
- in
- interest
- involved
- Leads
- leveraging
- Localization
- Long
- material
- mechanism
- Mitochondria
- modified
- molecule
- more
- movement
- mRNA
- multiple
- next
- number
- ONE
- open
- Other
- outside
- pairs
- plato
- Plato Data Intelligence
- PlatoData
- population
- positioned
- positioning
- present
- Prior
- probe
- produce
- Product
- properties
- protect
- Protein
- Proteins
- ranging
- recognized
- referred
- region
- replication
- resonance
- resulting
- RNA
- sensitive
- Sequence
- Shape
- Signal
- single
- Size
- small
- specific
- specifically
- specificity
- Stem
- structure
- studies
- such
- suitable
- Target
- The
- thereby
- thousands
- throughout
- to
- top
- transfer
- triggered
- under
- unique
- Variant
- which
- will
- zephyrnet